Even though this particular legend was designed with those needs, it should be simple to extrapolate from that to build legends based on other criteria.
Let's start by creating a map with a standard legend, and then we move on to customization later.
And now let's map the data:
# map data:
map("state", # base
col = "gray80",
fill = TRUE,
lty = 0)
map("state", # data
col = colcode,
fill = TRUE,
lty = 0,
add = TRUE)
map("state", # border
col = "gray",
lwd = 1.4,
lty = 1,
add = TRUE)
And finally let's add our default legend:
legend("bottomleft", # position
legend = names(attr(colcode, "table")),
title = "Percent",
fill = attr(colcode, "palette"),
cex = 0.56,
bty = "n") # border
Here's the output of this code (see
map-standard-legend.R in the gist):
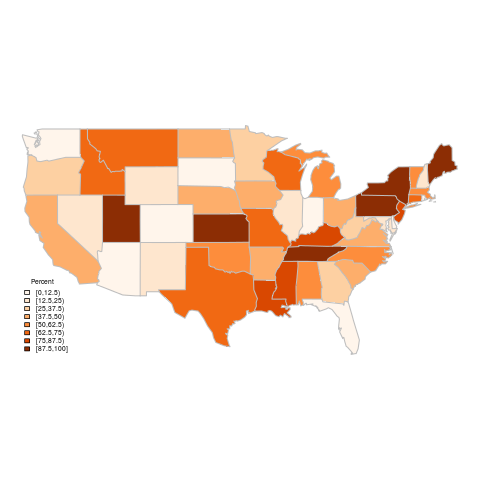 |
| Percent of power coming from coal sources (standard legend) |
Custom Legend
Next we want to add a few lines here and there to enhance the legend.
For starters, let's deal with NA values. We don't have any in this particular dataset, but if we did, we would have seen they were left as the base color of the map and not included in the legend.
After our former code setting up the colors, we should add the color for NAs. It's important that these lines go after all the other set up code, or the wrong colors will be mapped.
# set up colors:
plotclr <- brewer.pal(nclr, "Oranges")
plotvar <- state$coal
class <- classIntervals(plotvar,
nclr,
style = "fixed",
fixedBreaks = seq(min, max, breaks))
colcode <- findColours(class,
plotclr)
NAColor <- "gray80"
plotclr <- c(plotclr, NAColor)
We also want to let the map know to have our NA color as the default color, so the map will use that instead of having those areas be transparent:
# map data:
map("state", # base
col = NAColor,
fill = TRUE,
lty = 0)
map("state", # data
col = colcode,
fill = TRUE,
lty = 0,
add = TRUE)
map("state", # border
col = "gray",
lwd = 1.4,
lty = 1,
add = TRUE)
Next, we want to set up the legend text. For all but the last interval, we want it to say
i ≤ n < (i + breaks). The last interval should be
i ≤ n ≤ (i + breaks).
This can be accomplished by
# set legend text:
legendText <- c()
for(i in seq(min, max - (max - min) / nclr, (max - min) / nclr)) {
if (i == max(seq(min, max - (max - min) / nclr, (max - min) / nclr))) {
legendText <- c(legendText, paste(round(i,3), "\u2264 n \u2264", round(i + (max - min) / nclr,3)))
} else
legendText <- c(legendText, paste(round(i,3), "\u2264 n <", round(i + (max - min) / nclr,3)))
}
But we also want to include NAs in the legend, so we need to add a line:
# set legend text:
legendText <- c()
for(i in seq(min, max - (max - min) / nclr, (max - min) / nclr)) {
if (i == max(seq(min, max - (max - min) / nclr, (max - min) / nclr))) {
legendText <- c(legendText, paste(round(i,3), "\u2264 n \u2264", round(i + (max - min) / nclr,3)))
if (!is.na(NAColor)) legendText <- c(legendText, "NA")
} else
legendText <- c(legendText, paste(round(i,3), "\u2264 n <", round(i + (max - min) / nclr,3)))
}
And finally we need to add the legend to the map:
legend("bottomleft", # position
legend = legendText,
title = "Percent",
fill = plotclr,
cex = 0.56,
bty = "n") # border
The new map (see
map-new-legend.R) meets all the criteria we started with that the original legend didn't have.
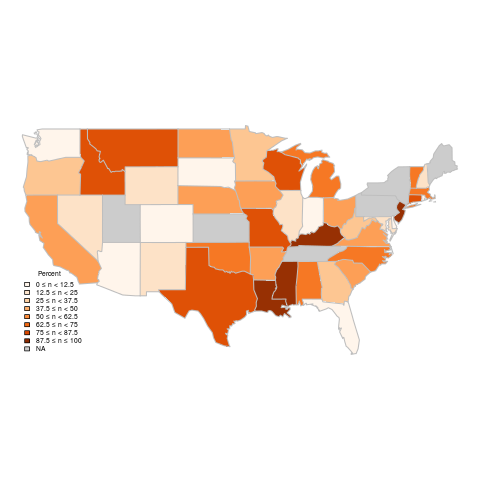 |
| Percent of power coming from coal sources (custom legend) |
Code is available in a
gist.